class: center, middle, inverse, title-slide # 一元配置の分散分析と多重比較 ## 『Rで学ぶ統計学入門』第6章 ### 2018-07-09 --- class: middle * 3標本の平均値を比較する場合に `\(t\)` 検定を繰り返すのは誤り * 分散分析と多重比較法を用いる --- ## 今日使うパッケージ ```r # library(multcomp) library(dplyr) ``` ``` ## ## Attaching package: 'dplyr' ``` ``` ## The following objects are masked from 'package:stats': ## ## filter, lag ``` ``` ## The following objects are masked from 'package:base': ## ## intersect, setdiff, setequal, union ``` ```r library(tidyr) library(ggplot2) library(readr) ``` --- ## _t_ 検定を繰り返すのは誤り * 3つの標本 A,B,C の平均値 `\(\bar{X}_\mathrm{A}\)`, `\(\bar{X}_\mathrm{B}\)`, `\(\bar{X}_\mathrm{C}\)` を考える * これらが同一母集団から得られた標本かどうかを帰無仮説検定する * 帰無仮説 H<sub>0</sub>: `$$\mu_\mathrm{A} = \mu_\mathrm{B} = \mu_\mathrm{C}$$` -- * ペアごとに有意水準5%で `\(t\)` 検定をすると,3回で正しく H<sub>0</sub>を棄却できる確率は, `$$(1-0.95)^3=0.95^3=0.857$$` となり,第1種過誤の確率は, `$$1-0.857= 0.143$$` となってしまう --- class: center, middle, inverse ## 一元配置分散分析(1-way ANOVA) の原理 --- ### 分散分析(Analysis of variance, ANOVA) 「観測データにおける変動を誤差変動と各要因およびそれらの交互作用による変動に分解することによって、要因および交互作用の効果を判定する、統計的仮説検定の一手法」(Wikipedia) #### 前提 * 正規性: 各群の母集団分布は正規分布に従う * 等分散: すべての群を通して母集団は等しい #### ばらつきの要因の分解 * データのばらつき: 処理 + 偶然 * 群間: 処理 + 偶然 * 群内: 偶然 * 群間分散と群内分散の比から `\(F\)` 値 を求め, `\(F\)` 分布を用いて帰無仮説検定する `$$F = \frac{\text{群間分散}}{\text{群内分散}} = \frac{\text{処理 + 偶然}}{偶然}$$` --- ### 1-way ANOVA の原理 3つの標本があるとする .pull-left[ * 確かめたいこと: 3つの標本が同一母集団から抽出されたかどうか * 帰無仮説: `\(\mu_\mathrm{A} = \mu_\mathrm{B} = \mu_\mathrm{C}\)` * 対立仮説: `\(\overline{\mu_\mathrm{A} = \mu_\mathrm{B} = \mu_\mathrm{C}}\)` * 帰無仮説が棄却された `\(\rightarrow\)` 少なくとも一つに有意差あり * どの平均値間に有意差があるかは __多重比較法__ ] .pull-right[ | 群 | 標本サイズ | データ | 平均 | |:---:|:---:|:---:|:---:| | 1 | `\(n_1\)` | `\(X_{1 1}\)`, `\(X_{1 2}\)`, `\(\cdots\)`, `\(X_{1 n_1 }\)` | `\(\bar{X}_1\)` | | 2 | `\(n_2\)` | `\(X_{2 1}\)`, `\(X_{2 2}\)`, `\(\cdots\)`, `\(X_{2 n_2 }\)` | `\(\bar{X}_2\)` | | `\(\vdots\)` | `\(\vdots\)` | `\(\vdots\)` | `\(\vdots\)` | | `\(i\)` | `\(n_i\)` | `\(X_{i 1}\)`, `\(X_{i 2}\)`, `\(\cdots\)`, `\(X_{i n_i }\)` | `\(\bar{X}_i\)` | | `\(\vdots\)` | `\(\vdots\)` | `\(\vdots\)` | `\(\vdots\)` | | `\(m\)` | `\(n_m\)` | `\(X_{m 1}\)`, `\(X_{m 2}\)`, `\(\cdots\)`, `\(X_{m n_m }\)` | `\(\bar{X}_m\)` | `$$N=n_1 + n_2 + \cdots + n_i + \cdots + n_m$$` ] --- ### 1-way ANOVA の原理 平方和から分散を求める .pull-left[ * 総平均を `\(\bar{X}\)` とすると,全体偏差は `\(X_{ij} - \bar{X}\)` * 全体偏差は群内偏差 `\(X_{ij} - \bar{X}_i\)` と 群間偏差 `\(\bar{X}_i - \bar{X}\)` に分割できる `$$X_{ij} - \bar{X} = (X_{ij} - \bar{X}_i) + (\bar{X}_i - \bar{X})$$` * 総平方和 `\(SS_T\)`は, `$$SS_\mathrm{T} = \sum_{i=1}^{m}\sum_{j=1}^{n_i}(X_{ij} - \bar{X})^2$$` ] .pull-right[ | 群 | 標本サイズ | データ | 平均 | |:---:|:---:|:---:|:---:| | 1 | `\(n_1\)` | `\(X_{1 1}\)`, `\(X_{1 2}\)`, `\(\cdots\)`, `\(X_{1 n_1 }\)` | `\(\bar{X}_1\)` | | 2 | `\(n_2\)` | `\(X_{2 1}\)`, `\(X_{2 2}\)`, `\(\cdots\)`, `\(X_{2 n_2 }\)` | `\(\bar{X}_2\)` | | `\(\vdots\)` | `\(\vdots\)` | `\(\vdots\)` | `\(\vdots\)` | | `\(i\)` | `\(n_i\)` | `\(X_{i 1}\)`, `\(X_{i 2}\)`, `\(\cdots\)`, `\(X_{i n_i }\)` | `\(\bar{X}_i\)` | | `\(\vdots\)` | `\(\vdots\)` | `\(\vdots\)` | `\(\vdots\)` | | `\(m\)` | `\(n_m\)` | `\(X_{m 1}\)`, `\(X_{m 2}\)`, `\(\cdots\)`, `\(X_{m n_m }\)` | `\(\bar{X}_m\)` | `$$N=n_1 + n_2 + \cdots + n_i + \cdots + n_m$$` `$$\bar{X}=\frac{1}{N}\sum_{i=1}^{m}\sum_{j=1}^{n_i}X_{ij}$$` ] --- ### 1-way ANOVA の原理 全体偏差を群内偏差と群間偏差に分割して, .pull-left[ `$$SS_\mathrm{T} = \sum_{i=1}^{m}\sum_{j=1}^{n_i}\{(X_{ij} - \bar{X}_i) + (\bar{X}_i - \bar{X})\}^2$$` 展開して整理すると, `$$SS_\mathrm{T} = \sum_{i=1}^{m}\sum_{j=1}^{n_i}(X_{ij} - \bar{X}_i)^2 + \sum_{i=1}^{m}n_i(\bar{X}_i - \bar{X})^2$$` となり,総平方和は * 群内平方和 `\(SS_\mathrm{W} = \sum_{i=1}^{m}\sum_{j=1}^{n_i}(X_{ij} - \bar{X}_i)^2\)` * 群間平方和 `\(SS_\mathrm{B} = \sum_{i=1}^{m}n_i(\bar{X}_i - \bar{X})^2\)` に分割される ] .pull-right[ | 群 | 標本サイズ | データ | 平均 | |:---:|:---:|:---:|:---:| | 1 | `\(n_1\)` | `\(X_{1 1}\)`, `\(X_{1 2}\)`, `\(\cdots\)`, `\(X_{1 n_1 }\)` | `\(\bar{X}_1\)` | | 2 | `\(n_2\)` | `\(X_{2 1}\)`, `\(X_{2 2}\)`, `\(\cdots\)`, `\(X_{2 n_2 }\)` | `\(\bar{X}_2\)` | | `\(\vdots\)` | `\(\vdots\)` | `\(\vdots\)` | `\(\vdots\)` | | `\(i\)` | `\(n_i\)` | `\(X_{i 1}\)`, `\(X_{i 2}\)`, `\(\cdots\)`, `\(X_{i n_i }\)` | `\(\bar{X}_i\)` | | `\(\vdots\)` | `\(\vdots\)` | `\(\vdots\)` | `\(\vdots\)` | | `\(m\)` | `\(n_m\)` | `\(X_{m 1}\)`, `\(X_{m 2}\)`, `\(\cdots\)`, `\(X_{m n_m }\)` | `\(\bar{X}_m\)` | `$$N=n_1 + n_2 + \cdots + n_i + \cdots + n_m$$` `$$\bar{X}=\frac{1}{N}\sum_{i=1}^{m}\sum_{j=1}^{n_i}X_{ij}$$` ] --- ### 1-way ANOVA の原理 .pull-left[ 分散は平方和を自由度で割ると求められるので, * 群間分散 `\(MS_\mathrm{B} = SS_\mathrm{B} / (m - 1)\)` * 群間自由度 `\(df_\mathrm{B} = m - 1\)` * 群内分散 `\(MS_\mathrm{W} = SS_\mathrm{W} / m(n-1)\)` * 群内自由度 `\(df_\mathrm{W} = m(n - 1)\)` 統計量 `\(F\)` は, `$$F = \frac{MS_\mathrm{B}}{MS_\mathrm{W}}$$` で,これが自由度 ( `\(df_\mathrm{B}\)`, `\(df_\mathrm{W}\)` ) の `\(F\)` 分布に従うので, 棄却域に入るかどうかをみる 群間のばらつきが相対的に郡内のばらつきより大きいと `\(F\)` 値は大きくなる ] .pull-right[ <!-- --> `\(df=(3, 44)\)` の `\(F\)` 分布と, `\(\alpha=0.05\)` (累積確率95%) の位置 ] --- class: center, middle, inverse ## Rを使ったANOVAの事例 --- ### 五つの湖のストロンチウム濃度 データ読み込み ```r d.lake <- readr::read_csv("../../samplecode/Rで学ぶ統計学入門図版作成用(改訂版)/付録/table6-3.csv") ``` ``` ## Parsed with column specification: ## cols( ## stron = col_double(), ## lake = col_integer() ## ) ``` ```r glimpse(d.lake) ``` ``` ## Observations: 30 ## Variables: 2 ## $ stron <dbl> 28.2, 33.2, 36.4, 34.6, 29.1, 31.0, 39.6, 40.8, 37.9, 37... ## $ lake <int> 1, 1, 1, 1, 1, 1, 2, 2, 2, 2, 2, 2, 3, 3, 3, 3, 3, 3, 4,... ``` --- ### R で 1-way ANOVA * 要因: `lake` (5群) * 統計量 `\(F = 56.16\)` * 自由度 `(4, 25)` の `\(F\)` 分布 * `\(p < 0.05\)` で有意 ```r d.lake %>% mutate(lake = as.factor(lake)) %>% aov(stron ~ lake, data=.) %>% summary() ``` ``` ## Df Sum Sq Mean Sq F value Pr(>F) ## lake 4 2193.4 548.4 56.16 3.95e-12 *** ## Residuals 25 244.1 9.8 ## --- ## Signif. codes: 0 '***' 0.001 '**' 0.01 '*' 0.05 '.' 0.1 ' ' 1 ``` --- ### 計算過程(別解) .pull-left[ ```r # 群数 m <- 5 # 標本サイズ n <- 6 # 群内平方和 (SS.W <- d.lake %>% group_by(lake) %>% summarise(x = sum((stron - mean(stron))^2)) %>% summarise(SS.W = sum(x)) %>% .[["SS.W"]]) ``` ``` ## [1] 244.13 ``` ```r # 群間平方和 (SS.B <- d.lake %>% group_by(lake) %>% summarise(x=mean(stron)) %>% mutate(y = (x - mean(x))^2) %>% summarise(SS.B = n * sum(y)) %>% .[["SS.B"]] ) ``` ``` ## [1] 2193.442 ``` ] .pull-right[ ```r d.lake %>% mutate(lake = as.factor(lake)) %>% aov(stron ~ lake, data=.) %>% summary() ``` ``` ## Df Sum Sq Mean Sq F value Pr(>F) ## lake 4 2193.4 548.4 56.16 3.95e-12 *** ## Residuals 25 244.1 9.8 ## --- ## Signif. codes: 0 '***' 0.001 '**' 0.01 '*' 0.05 '.' 0.1 ' ' 1 ``` ] --- ### 計算過程(別解) .pull-left[ ```r # 群内自由度 (df.B <- m - 1) ``` ``` ## [1] 4 ``` ```r # 群内分散 (MS.B <- SS.B / df.B) ``` ``` ## [1] 548.3605 ``` ```r # 群間自由度 (df.W <- m * (n - 1)) ``` ``` ## [1] 25 ``` ```r # 群間分散 (MS.W <- SS.W / df.W) ``` ``` ## [1] 9.7652 ``` ] .pull-right[ ```r d.lake %>% mutate(lake = as.factor(lake)) %>% aov(stron ~ lake, data=.) %>% summary() ``` ``` ## Df Sum Sq Mean Sq F value Pr(>F) ## lake 4 2193.4 548.4 56.16 3.95e-12 *** ## Residuals 25 244.1 9.8 ## --- ## Signif. codes: 0 '***' 0.001 '**' 0.01 '*' 0.05 '.' 0.1 ' ' 1 ``` ] --- ### 計算過程(別解) .pull-left[ ```r # F値 (F.value <- MS.B / MS.W) ``` ``` ## [1] 56.15456 ``` ```r # p値 1 - pf(F.value, df.B, df.W) ``` ``` ## [1] 3.947953e-12 ``` ] .pull-right[ ```r d.lake %>% mutate(lake = as.factor(lake)) %>% aov(stron ~ lake, data=.) %>% summary() ``` ``` ## Df Sum Sq Mean Sq F value Pr(>F) ## lake 4 2193.4 548.4 56.16 3.95e-12 *** ## Residuals 25 244.1 9.8 ## --- ## Signif. codes: 0 '***' 0.001 '**' 0.01 '*' 0.05 '.' 0.1 ' ' 1 ``` ] ??? `F` は `FALSE` なので `F.value` にする --- ### ブタの体重増加への餌の種類の影響 .pull-left[ * ある品種のブタに4種類の餌を与えた * 160日後の体重(kg)のデータ * Pig 列にNA(欠損値) あり ```r d.pig <- read_csv("../../samplecode/Rで学ぶ統計学入門図版作成用(改訂版)/付録/table6-4.csv") glimpse(d.pig) ``` ``` ## Observations: 20 ## Variables: 2 ## $ Pig <dbl> 60.8, 57.0, 65.0, 58.6, 61.7, 68.7, 67.7, 74.0, 66.3, 69.... ## $ feed <int> 1, 1, 1, 1, 1, 2, 2, 2, 2, 2, 3, 3, 3, 3, 3, 4, 4, 4, 4, 4 ``` ```r d.pig %>% filter(is.na(Pig)) ``` ``` ## # A tibble: 1 x 2 ## Pig feed ## <dbl> <int> ## 1 NA 3 ``` ] .pull-right[ ```r d.pig %>% ggplot(aes(x=feed, y=Pig, group=feed)) + geom_boxplot() + theme_bw() + theme(panel.grid=element_blank()) ``` ``` ## Warning: Removed 1 rows containing non-finite values (stat_boxplot). ``` <!-- --> ] --- ### ブタの体重増加への餌の種類の影響 .pull-left[ * NA は無視されるので,そのまま ANOVA を実行 * `\(p < 0.05\)` で,エサの効果あり ```r d.pig %>% mutate(feed = as.factor(feed)) %>% aov(Pig ~ feed, data=.) %>% summary() ``` ``` ## Df Sum Sq Mean Sq F value Pr(>F) ## feed 3 4226 1408.8 164.6 1.06e-11 *** ## Residuals 15 128 8.6 ## --- ## Signif. codes: 0 '***' 0.001 '**' 0.01 '*' 0.05 '.' 0.1 ' ' 1 ## 1 observation deleted due to missingness ``` ] .pull-right[ ```r d.pig %>% ggplot(aes(x=feed, y=Pig, group=feed)) + geom_boxplot() + theme_bw() + theme(panel.grid=element_blank()) ``` ``` ## Warning: Removed 1 rows containing non-finite values (stat_boxplot). ``` <!-- --> ] --- ### チューキー(Tukey)のHSD検定 .pull-left[ * 残差 `\(MS\)` ( `\(MS_\mathrm{W}\)` )から標準誤差( `\(SE\)` )を求める `$$SE = \sqrt{MS_\mathrm{W}/n}$$` * 統計量 `\(q\)` を全てのペアについて求める * `\({}_5\mathrm{C}_2 = 10\)` 通り `$$|q| = \frac{\bar{X}_i - \bar{X}_j}{SE}$$` * `\(q\)` 棄の却率値を スチューデント化された `\(q\)`分布 から求める ] .pull-right[ ```r d.lake %>% mutate(lake = as.factor(lake)) %>% aov(stron ~ lake, data=.) %>% TukeyHSD() ``` ``` ## Tukey multiple comparisons of means ## 95% family-wise confidence level ## ## Fit: aov(formula = stron ~ lake, data = .) ## ## $lake ## diff lwr upr p adj ## 2-1 8.1500000 2.851355 13.448645 0.0011293 ## 3-1 12.0000000 6.701355 17.298645 0.0000053 ## 4-1 9.0166667 3.718021 14.315312 0.0003339 ## 5-1 26.2166667 20.918021 31.515312 0.0000000 ## 3-2 3.8500000 -1.448645 9.148645 0.2376217 ## 4-2 0.8666667 -4.431979 6.165312 0.9884803 ## 5-2 18.0666667 12.768021 23.365312 0.0000000 ## 4-3 -2.9833333 -8.281979 2.315312 0.4791100 ## 5-3 14.2166667 8.918021 19.515312 0.0000003 ## 5-4 17.2000000 11.901355 22.498645 0.0000000 ``` ] --- ### チューキー(Tukey)のHSD検定 .pull-left[ <!-- --> ] .pull-right[ ```r d.lake %>% mutate(lake = as.factor(lake)) %>% aov(stron ~ lake, data=.) %>% TukeyHSD() ``` ``` ## Tukey multiple comparisons of means ## 95% family-wise confidence level ## ## Fit: aov(formula = stron ~ lake, data = .) ## ## $lake ## diff lwr upr p adj ## 2-1 8.1500000 2.851355 13.448645 0.0011293 ## 3-1 12.0000000 6.701355 17.298645 0.0000053 ## 4-1 9.0166667 3.718021 14.315312 0.0003339 ## 5-1 26.2166667 20.918021 31.515312 0.0000000 ## 3-2 3.8500000 -1.448645 9.148645 0.2376217 ## 4-2 0.8666667 -4.431979 6.165312 0.9884803 ## 5-2 18.0666667 12.768021 23.365312 0.0000000 ## 4-3 -2.9833333 -8.281979 2.315312 0.4791100 ## 5-3 14.2166667 8.918021 19.515312 0.0000003 ## 5-4 17.2000000 11.901355 22.498645 0.0000000 ``` ] --- class: center, middle, inverse ## 多重比較とは --- ### 多重比較とは * 3群以上の平均値が得られた時にどの平均値間に差があるかを検定する * `\(t\)` 検定を繰り返すと有意水準が保たれないので,これを調整する * 事前比較: あらかじめどの群間に差があるか分かっている `\(\rightarrow\)` ANOVAなしで多重比較 * 事後比較: どの群間に差があるか分かってない `\(\rightarrow\)` ANOVA してから多重比較 方法には例えば, 1. Tukey-Kramer 1. Dunnett 1. Scheffé 1. Wiliams などがある --- ### 多重比較法(1)チューキーとクレーマーの方法 .pull-left[ * 群間で全ての対比較 * 例: グループごとの点数 ```r d_67 <- read_csv("../../samplecode/Rで学ぶ統計学入門図版作成用(改訂版)/付録/table6-7.csv") glimpse(d_67) ``` ``` ## Observations: 30 ## Variables: 2 ## $ score <int> 15, 16, 17, 15, 19, 19, 17, 14, 17, 16, 15, 17, 16, 18, ... ## $ group <chr> "g1", "g1", "g1", "g1", "g1", "g1", "g1", "g2", "g2", "g... ``` ```r res.d_67 <- d_67 %>% mutate(group = as.factor(group)) %>% aov(score ~ group, data=.) res.d_67 %>% summary() ``` ``` ## Df Sum Sq Mean Sq F value Pr(>F) ## group 3 80.28 26.759 10.61 9.81e-05 *** ## Residuals 26 65.59 2.523 ## --- ## Signif. codes: 0 '***' 0.001 '**' 0.01 '*' 0.05 '.' 0.1 ' ' 1 ``` ] .pull-right[ ```r gp.d_67 <- ggplot(d_67, aes(x=group, y=score, group=group)) + geom_boxplot() + theme_bw() + theme(panel.grid=element_blank()) gp.d_67 ``` <!-- --> ] --- ### 多重比較法(1)チューキーとクレーマーの方法 .pull-left[ ```r res.d_67 %>% TukeyHSD() ``` ``` ## Tukey multiple comparisons of means ## 95% family-wise confidence level ## ## Fit: aov(formula = score ~ group, data = .) ## ## $group ## diff lwr upr p adj ## g2-g1 -0.7142857 -3.0433003 1.614729 0.8342852 ## g3-g1 1.6428571 -0.6122015 3.897916 0.2143839 ## g4-g1 3.5178571 1.2627985 5.772916 0.0012127 ## g3-g2 2.3571429 0.1020842 4.612201 0.0380588 ## g4-g2 4.2321429 1.9770842 6.487201 0.0001269 ## g4-g3 1.8750000 -0.3035936 4.053594 0.1102113 ``` ] .pull-right[ ```r gp.d_67 + annotate(geom="text", label=c("ab", "a", "bc", "c"), x=c(1, 2, 3, 4)+0.1, y=23) ``` <!-- --> ] --- ### 多重比較法(2): ダネットの方法 .pull-left[ * 対象群と処理群の対比較 * 例: 対象群C と処理群g2,g3,g4 の比較 ```r d_68 <- read_csv("../../samplecode/Rで学ぶ統計学入門図版作成用(改訂版)/付録/table6-8.csv") glimpse(d_68) ``` ``` ## Observations: 34 ## Variables: 2 ## $ score <int> 7, 9, 8, 6, 9, 8, 11, 10, 8, 8, 8, 9, 10, 8, 9, 9, 10, 1... ## $ group <chr> "C", "C", "C", "C", "C", "C", "C", "C", "C", "C", "g2", ... ``` ```r res.d_68 <- d_68 %>% mutate(group=as.factor(group)) %>% aov(score ~ group, data=.) res.d_68 %>% summary() ``` ``` ## Df Sum Sq Mean Sq F value Pr(>F) ## group 3 64.73 21.577 12.18 2.2e-05 *** ## Residuals 30 53.15 1.772 ## --- ## Signif. codes: 0 '***' 0.001 '**' 0.01 '*' 0.05 '.' 0.1 ' ' 1 ``` ] .pull-right[ ```r gp.d_68 <- ggplot(d_68, aes(x=group, y=score, group=group)) + geom_boxplot() + theme_bw() + theme(panel.grid=element_blank()) gp.d_68 ``` <!-- --> ] --- ### 多重比較法(2): ダネットの方法 .pull-left[ * group 3, 4 と コントロール間で有意差あり ```r multcomp::glht(res.d_68, linfct = multcomp::mcp(group="Dunnett")) %>% summary() ``` ``` ## ## Simultaneous Tests for General Linear Hypotheses ## ## Multiple Comparisons of Means: Dunnett Contrasts ## ## ## Fit: aov(formula = score ~ group, data = .) ## ## Linear Hypotheses: ## Estimate Std. Error t value Pr(>|t|) ## g2 - C == 0 0.9750 0.6314 1.544 0.309 ## g3 - C == 0 2.6000 0.6314 4.118 <0.001 *** ## g4 - C == 0 3.4750 0.6314 5.504 <0.001 *** ## --- ## Signif. codes: 0 '***' 0.001 '**' 0.01 '*' 0.05 '.' 0.1 ' ' 1 ## (Adjusted p values reported -- single-step method) ``` ] .pull-right[ ```r gp.d_68 ``` <!-- --> ] --- ### シーケンシャル・ボンフェローニの方法(ホルムの方法) .pull-left[ * 帰無仮説の数に応じて有意水準を調整する * `\(p\)` 値の小さい順に帰無仮説を並べ替える * 残りの帰無仮説の数に応じて有意水準は調整される 例: 表6.7 ```r d_67 %>% {pairwise.t.test(.$score, .$group, p.adj="holm")} ``` ``` ## ## Pairwise comparisons using t tests with pooled SD ## ## data: .$score and .$group ## ## g1 g2 g3 ## g2 0.40782 - - ## g3 0.11243 0.03239 - ## g4 0.00112 0.00014 0.07800 ## ## P value adjustment method: holm ``` ] .pull-right[ 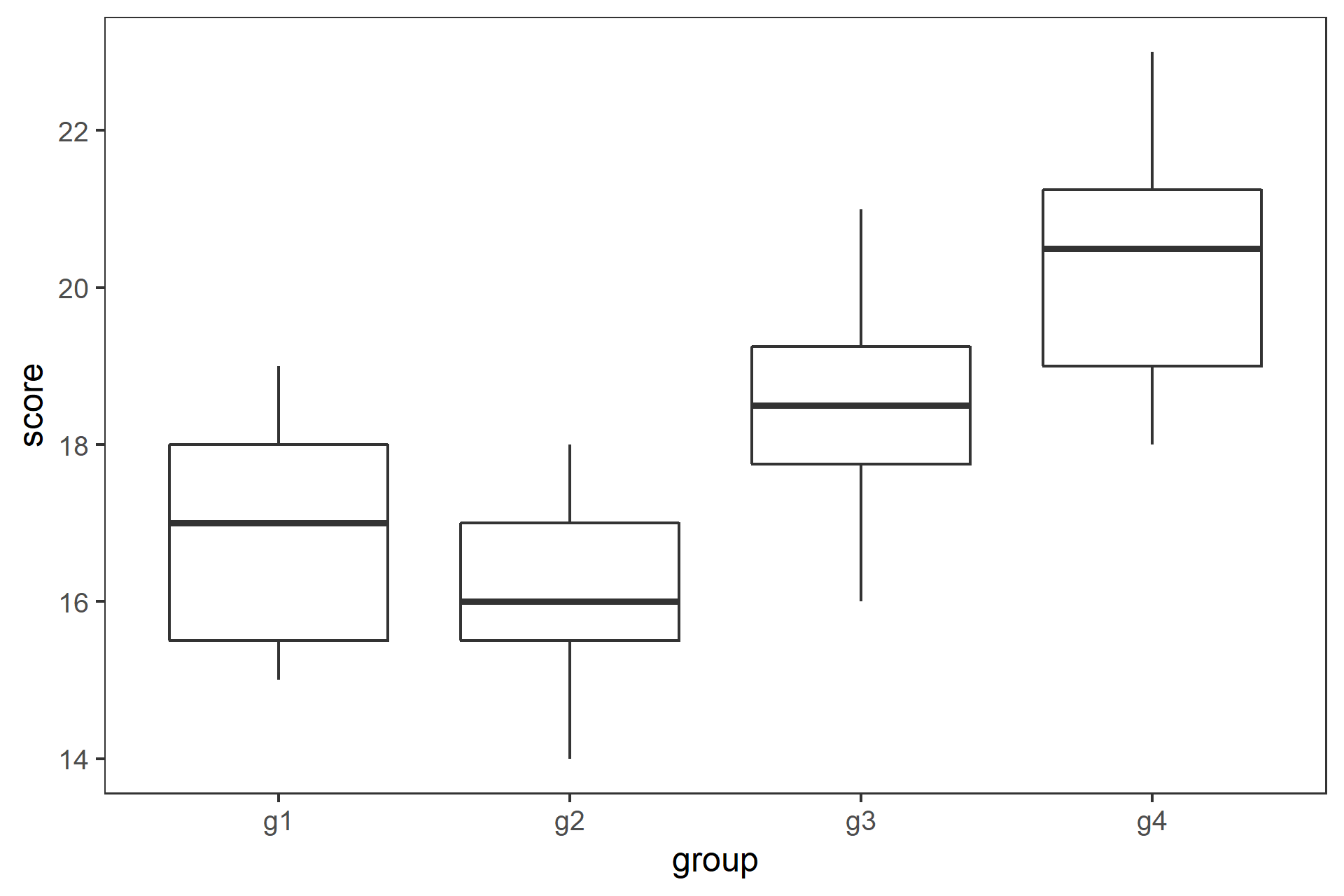<!-- --> ] --- ### シーケンシャル・ボンフェローニの方法(ホルムの方法) .pull-left[ 例: 表6.9 ```r d_69 <- read_csv("../../samplecode/Rで学ぶ統計学入門図版作成用(改訂版)/付録/table6-9.csv") glimpse(d_69) ``` ``` ## Observations: 22 ## Variables: 2 ## $ score <int> 58, 52, 72, 60, 67, 62, 53, 74, 69, 60, 65, 62, 69, 75, ... ## $ group <chr> "g1", "g1", "g1", "g1", "g1", "g2", "g2", "g2", "g2", "g... ``` ```r d_69 %>% {pairwise.t.test(.$score, .$group, p.adj="holm")} ``` ``` ## ## Pairwise comparisons using t tests with pooled SD ## ## data: .$score and .$group ## ## g1 g2 g3 ## g2 0.7169 - - ## g3 0.1482 0.1894 - ## g4 0.0037 0.0072 0.1894 ## ## P value adjustment method: holm ``` ] .pull-right[ ```r ggplot(d_69, aes(x=group, y=score, group=group)) + geom_boxplot() + theme_bw() + theme(panel.grid=element_blank()) + annotate(geom="text", label=c("a", "a", "ab", "b"), x=c(1, 2, 3, 4)+0.1, y=88) ``` 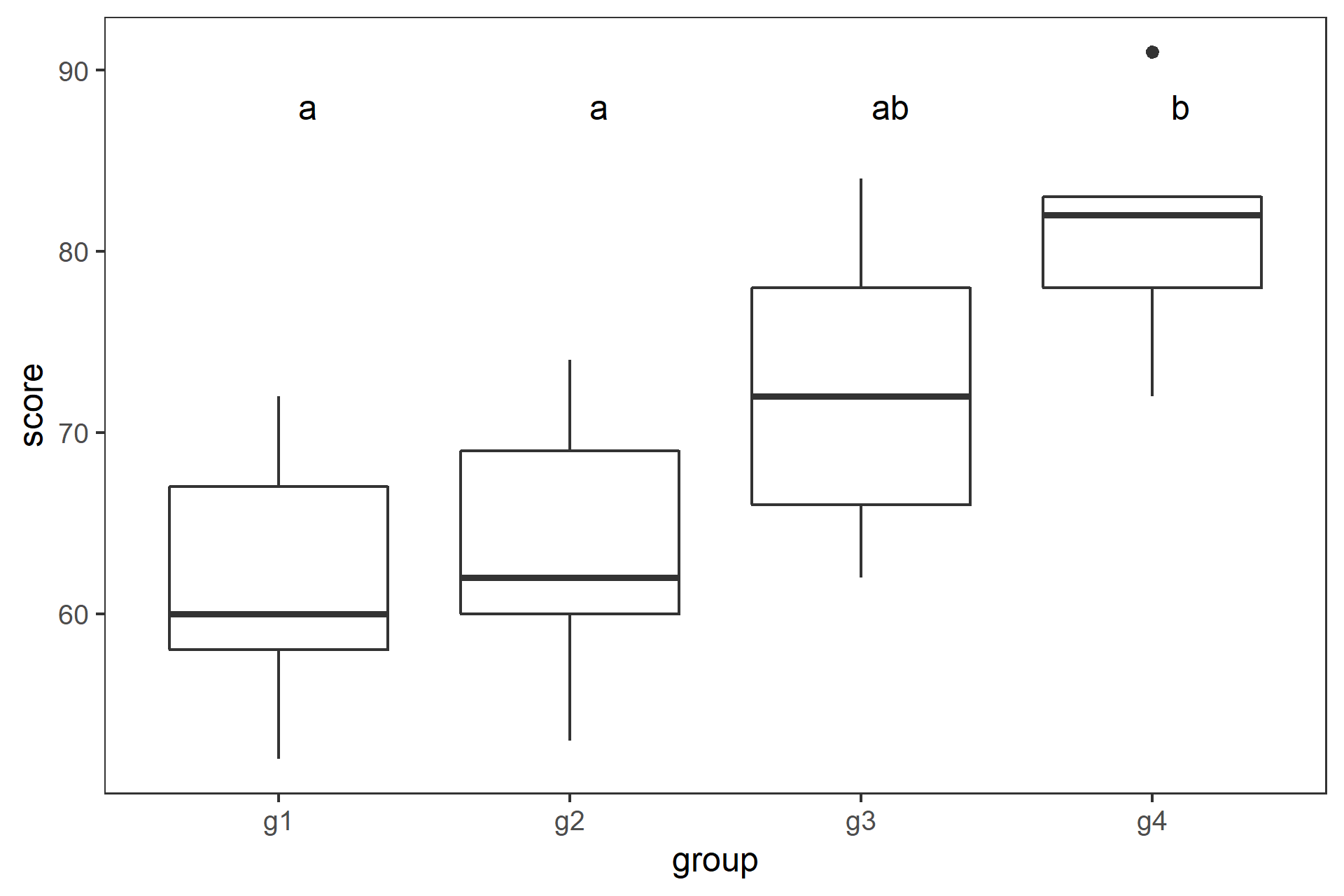<!-- --> ] --- ## まとめ * 3群以上の比較は `\(t\)` 検定ではなく分散分析を使う * 分散分析では処理効果の有無が判定できる * どの群に有意差があるのかを判定するときには多重比較法を使う * 多重比較法には様々な種類があるので,データの種類に応じて適切なものを用いる * 現代では使わない方がよいとされているものもあるので注意する --- ## 演習問題1:表6.4 ブタデータ .pull-left[ ```r d.pig %>% mutate(feed=as.factor(feed)) %>% summary() ``` ``` ## Pig feed ## Min. : 57.00 1:5 ## 1st Qu.: 65.65 2:5 ## Median : 74.00 3:5 ## Mean : 78.01 4:5 ## 3rd Qu.: 89.10 ## Max. :102.60 ## NA's :1 ``` ```r d.pig %>% group_by(feed) %>% summarise(mean=mean(Pig, na.rm=TRUE), sd=sd(Pig, na.rm=TRUE)) ``` ``` ## # A tibble: 4 x 3 ## feed mean sd ## <int> <dbl> <dbl> ## 1 1 60.6 3.06 ## 2 2 69.3 2.93 ## 3 3 100. 2.77 ## 4 4 86.2 2.90 ``` ] .pull-right[ ```r d.pig %>% ggplot(aes(x=feed, y=Pig, group=feed)) + geom_boxplot() + theme_bw() + theme(panel.grid=element_blank()) ``` ``` ## Warning: Removed 1 rows containing non-finite values (stat_boxplot). ``` 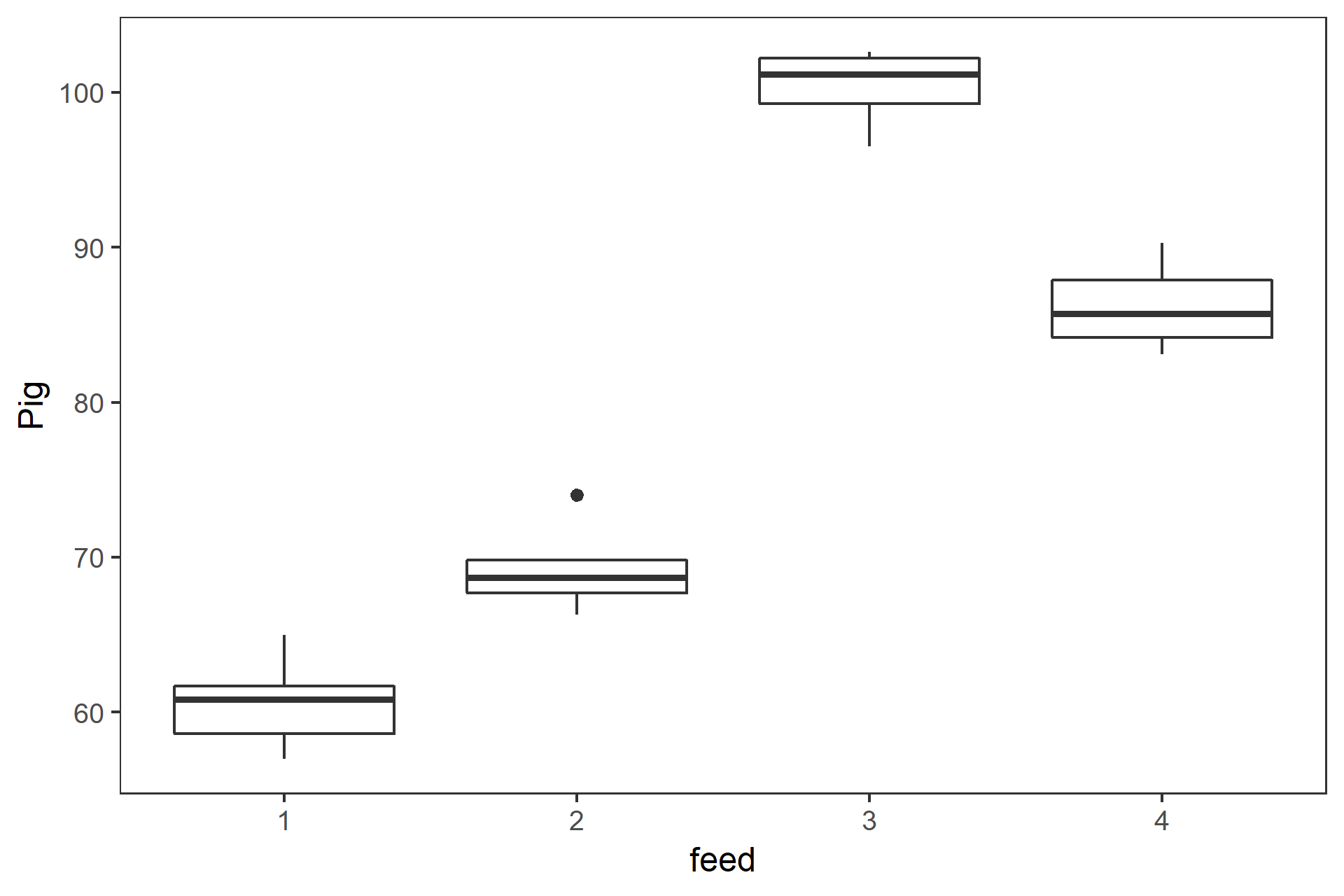<!-- --> ] .pull-left[ チューキーHSD ```r d.pig %>% mutate(feed=as.factor(feed)) %>% aov(Pig ~ feed, data=.) %>% TukeyHSD() ``` ``` ## Tukey multiple comparisons of means ## 95% family-wise confidence level ## ## Fit: aov(formula = Pig ~ feed, data = .) ## ## $feed ## diff lwr upr p adj ## 2-1 8.68 3.347895 14.012105 0.0014725 ## 3-1 39.73 34.074449 45.385551 0.0000000 ## 4-1 25.62 20.287895 30.952105 0.0000000 ## 3-2 31.05 25.394449 36.705551 0.0000000 ## 4-2 16.94 11.607895 22.272105 0.0000009 ## 4-3 -14.11 -19.765551 -8.454449 0.0000168 ``` ] .pull-right[ ホルム ```r d.pig %>% {pairwise.t.test(.$Pig, .$feed, p.adj= "holm")} ``` ``` ## ## Pairwise comparisons using t tests with pooled SD ## ## data: .$Pig and .$feed ## ## 1 2 3 ## 2 0.00029 - - ## 3 1.6e-11 4.6e-10 - ## 4 2.4e-09 4.7e-07 6.2e-06 ## ## P value adjustment method: holm ``` ] --- ## 演習問題2:表6.9 投薬データ(g1 が対象群) .pull-left[ ```r d_69 %>% mutate(group=as.factor(group)) %>% summary() ``` ``` ## score group ## Min. :52.00 g1:5 ## 1st Qu.:62.00 g2:5 ## Median :70.50 g3:6 ## Mean :70.36 g4:6 ## 3rd Qu.:78.50 ## Max. :91.00 ``` ```r d_69 %>% group_by(group) %>% summarise(mean=mean(score), sd=sd(score)) ``` ``` ## # A tibble: 4 x 3 ## group mean sd ## <chr> <dbl> <dbl> ## 1 g1 61.8 7.82 ## 2 g2 63.6 8.14 ## 3 g3 72.3 8.48 ## 4 g4 81.2 6.40 ``` ] .pull-right[ ```r d_69 %>% ggplot(aes(x=group, y=score, group=group)) + geom_boxplot() + theme_bw() + theme(panel.grid = element_blank()) ``` 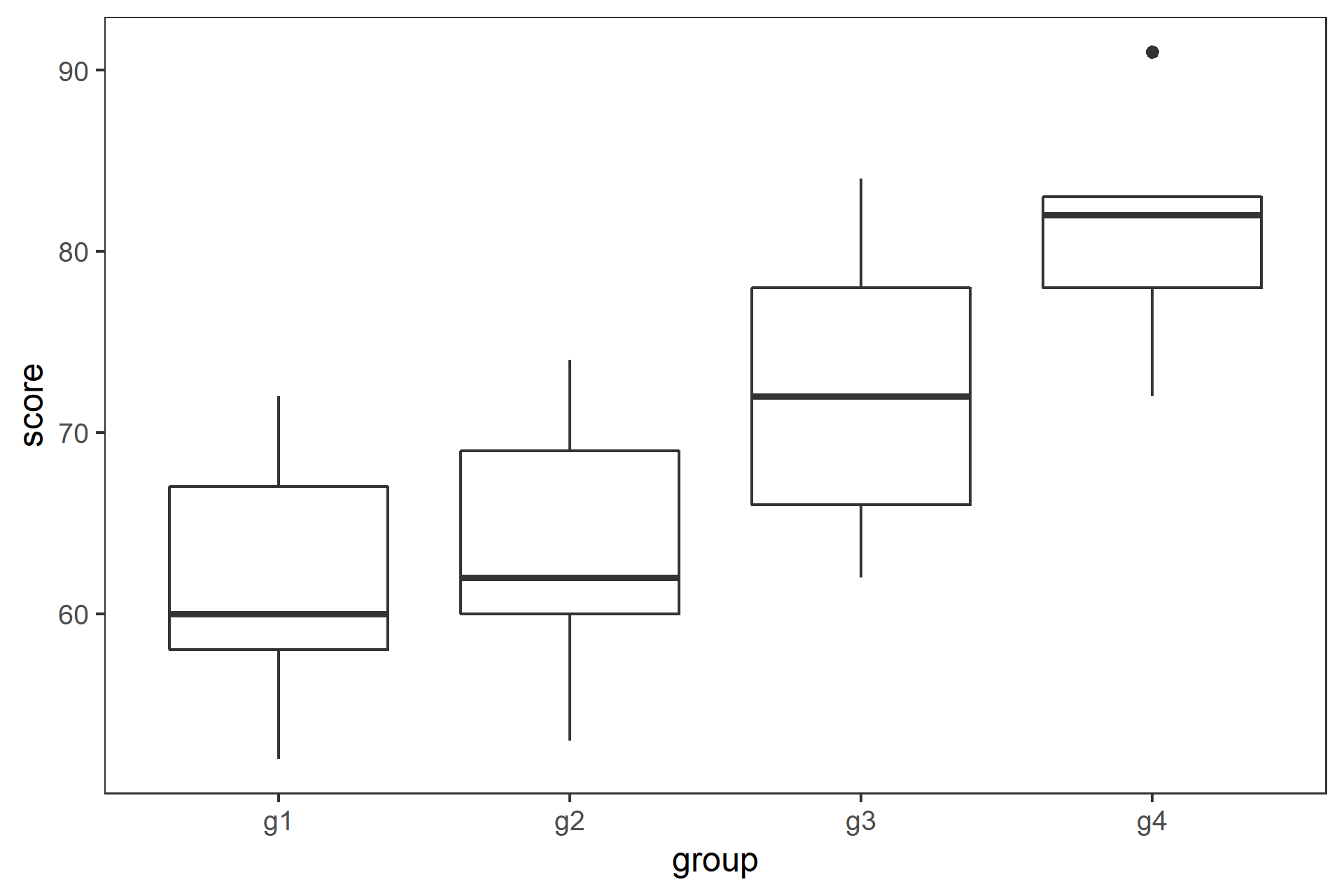<!-- --> ] --- ## 演習問題2 .pull-left[ ダネット ```r d_69 %>% mutate(group=as.factor(group)) %>% aov(score ~ group, data=.) %>% multcomp::glht(linfct = multcomp::mcp(group="Dunnet")) ``` ``` ## ## General Linear Hypotheses ## ## Multiple Comparisons of Means: Dunnett Contrasts ## ## ## Linear Hypotheses: ## Estimate ## g2 - g1 == 0 1.80 ## g3 - g1 == 0 10.53 ## g4 - g1 == 0 19.37 ``` ] .pull-right[ ホルム ```r d_69 %>% {pairwise.t.test(.$score, .$group, p.adj="holm")} ``` ``` ## ## Pairwise comparisons using t tests with pooled SD ## ## data: .$score and .$group ## ## g1 g2 g3 ## g2 0.7169 - - ## g3 0.1482 0.1894 - ## g4 0.0037 0.0072 0.1894 ## ## P value adjustment method: holm ``` ]